TriticeaeGenome

Improvement of Triticeae genomics
TriticeaeGenome project is a european EC 7th Framework Program (-Food, Agriculture and Fisheries) project, where INRA URGI platform is in partnership. TriticeaeGenome goal is to improve the genomics of Triticeae (wheat, barley). It is a large collaborative international project coordinated by C. Feuillet, INRA Clermont-Ferrand.
Links:
TriticeaeGenome project presentation
TriticeaeGenome collaborative workspace (project members only)
Project data summary:
| Data | Provider | Status | Public | Registered |
| Wheat | ||||
| Physical map 3B v1 and v2 | E. Paux (INRA GDEC, France) | done |
| |
| Physical map 1BL v1 and v2 | R. Philippe (INRA GDEC, France) | done |
| |
| Physical map 1AS v1 | J. Breen & T. Wicker (IPB, Zurich) | v1 |
| |
| Physical map 3DS v1 | J. Bartos & J. Dolozel (IEB, CZH) | v1 |
| |
| Physical map 1BS v1 | Z. Frenkel & A. Korol (Haifa, Israel) | v1 |
| |
| Genetic maps and markers : Neighbour3B, Neighbour1BL, SSR, ISBP. | P. Leroy & P. Sourdille (INRA GDEC, France) | done |
|
|
| QTL : TOR107 | M. Bogard & J. Le Gouis (INRA GDEC, France) | done |
| |
| QTL : CF9107xToisondorxQuebon | M. Bogard & J. Le Gouis (INRA GDEC, France) | done |
| |
| Barley | ||||
|
Genetic maps and markers : Stein consensus, Close Illumina consensus, Sato transcriptmap. | N. Stein (IPK, Germany) | done |
|
Project data submission:
For data submission and help, please contact us (michael.alaux[at]versailles.inra.fr, delphine.steinbach[at]versailles.inra.fr).
Duration: 01/01/2008 to 31/12/2011
Coordinator: Catherine Feuillet
URGI team: Michael Alaux, Daphné Verdelet, Delphine Steinbach, Hadi Quesneville
URGI contact: Michael Alaux (michael.alaux[at]versailles.inra.fr)
Description:
The project is divided into five research and technological workpackages, one workpackage for the dissemination and transfer of results and a workpackage dedicated to project management.
The two first WPs will develop genomic and genetic resources and assemble the knowledge required to produce physical maps of group 1 and 3 chromosomes in wheat and barley. These maps will be the basis for the isolation of genes of agronomic interest (WP3) that are located on these chromosomes and for which genetic resources are sufficiently developed to ensure candidate gene identification in the framework of the program. The markers that will be developed for anchoring the physical maps will enhance greatly the density of markers on the wheat, barley (and rye) genetic maps and provide state of the art tools for modern molecular breeding (WP4). Databases, software and web interfaces will be developed to assemble the data produced in the first four WPs and disseminate them to ensure their most efficient use and exploitation (WP5). The knowledge and tools generated by the project will be made available to the scientific and breeding communities as well as to the public (WP6) to ensure their best and most complete use for enhancing EU and international agricultural relevance and competitiveness.
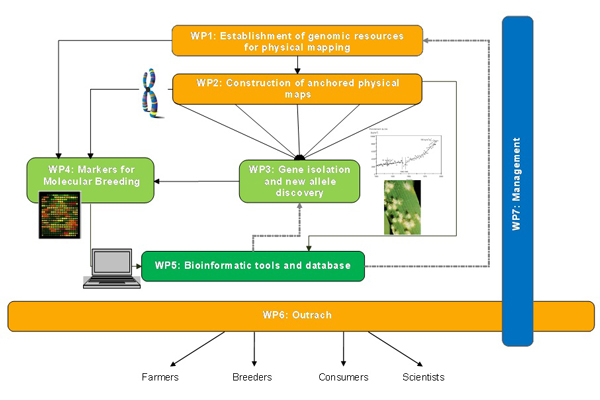
Detailed description of workpackages:
WP1 - Establishment of genomic resources for physical mapping of wheat
WP2 - Construction of anchored chromosome- and genomic physical maps of wheat and barley
WP3 - Gene isolation and new allele discovery
WP4 - Markers for molecular breeding
WP6 - Dissemination, Technology Transfer and training

