3BSeq

Sequencing, annotation and characterization of the bread wheat chromosome 3B
Data
URGI :
- Display 3B annotated pseudomolecule .
-
Download the 3B data
: Genomic DNA, CDSs, annotation of features and a README
ENA
: Sequences and annotations of the reference pseudomolecule and unassigned scaffolds have been deposited in ENA under the accession numbers HG670306 and CBUC010000001-CBUC010001450, respectively.
Publication
Choulet et al., Structural and Functional Partitioning of Bread Wheat Chromosome 3B
Science.
2014 Jul 18;345(6194):1249721. doi: 10.1126/science.1249721.
Abstract:
We produced a reference sequence of the 1-gigabase chromosome 3B of hexaploid bread wheat. By sequencing 8452 bacterial artificial chromosomes in pools, we assembled a sequence of 774 megabases carrying 5326 protein-coding genes, 1938 pseudogenes, and 85% of transposable elements. The distribution of structural and functional features along the chromosome revealed partitioning correlated with meiotic recombination. Comparative analyses indicated high wheat-specific inter- and intrachromosomal gene duplication activities that are potential sources of variability for adaption. In addition to providing a better understanding of the organization, function, and evolution of a large and polyploid genome, the availability of a high-quality sequence anchored to genetic maps will accelerate the identification of genes underlying important agronomic traits.
Project details
3BSeq is a flagship project funded by the ANR and France Agrimer for a duration of 3 years (2010-2013). The website and data from the 3BSEQ project are hosted by URGI together with all the other wheat resources.
The aim of 3BSeq is to sequence, annotate and characterization the bread wheat chromosome 3B. The project is coordinated by C. Feuillet, INRA Clermont-Ferrand.
3BSEQ is supported by the competitiveness cluster Céréales Vallée .
Duration: 01/01/2010 to 31/12/2012
Coordinator: Catherine Feuillet
Partner 1 : INRA GDEC (Clermont-Ferrand)
Frédéric Choulet, Etienne Paux, Pierre Sourdille, Philippe Leroy, Jérome Salse, Camille Rustenholz, Nicolas Guilhot, Catherine Feuillet (catherine.feuillet[at]clermont.inra.fr)
Partner 2: CEA Génoscope (Evry)
Julie Poulain, Adriana Alberti, Jean-Marc Aury, Arnaud Couloux, Patrick Wincker (pwincker[at]genoscope.cns.fr)
Partner 3: INRA URGI (Versailles)
Michael Alaux, Olivier Inizan, Erik Kimmel, Véronique Jamilloux, Isabelle Luyten, Sébastien Reboux, Delphine Steinbach, Hadi Quesneville (hadi.quesneville[at]versailles.inra.fr)
GDEC contact: Frédéric Choulet (frederic.choulet[at]clermont.inra.fr)
URGI contact: Michael Alaux (michael.alaux[at]versailles.inra.fr)
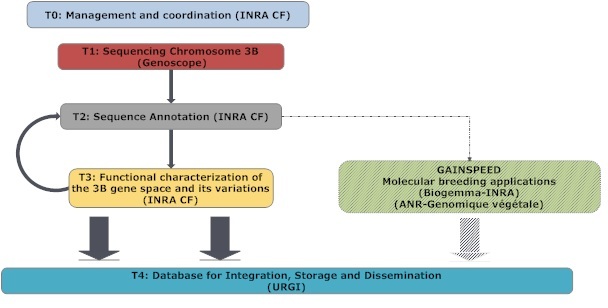
3BSEQ is organized in 4 scientific tasks and a management task (T0) represented below:
In 3BSEQ:
- T0 is focused on management of the project and aims at ensuring coordination between the partners, achievements of the objectives, dissemination of the results, and coordination at the international level with other projects within the International Wheat Genome Sequencing Consortium (IWGSC). Further, it will inform and organize annual meetings (at the PAG meeting every January) with the External advisory board. The Task is lead by INRA Clermont-Ferrand (Partner 1).
- T1 is dedicated to the sequencing of chromosome 3B and is led by Genoscope (Partner 2), the French National Sequencing Center . Genoscope will be in charge of developing the strategy, producing the sequence, and assembling it into scaffolds that represent chromosome 3B.
- T2 is led by Partner 1 and concerns the annotation of the sequence that will be produced in T1. Annotation will be done using a combination of automated tools already in place (TriAnnot pipeline and programs developed for the complete annotation of 20 Mb of contigs from chromosome 3B performed in the framework of an ANR Genoplante project ending this year (SMART, GPLA06001G) and for a project supported by Genoscope (AP2006). A workflow will be developed with URGI (Partner 3) to automatically retrieve, store and annotate the sequence as well as develop markers from the contig sequences as soon as they are released from Genoscope. Manual annotation will be performed also and specific target regions will be finished to obtain a complete contig sequence.
- T3 aims at establishing a transcriptional map of the chromosome 3B and at exploring the potential of CGH on tiling arrays to analyze copy number variations in wheat. Nimblegen tiling arrays that comprise gene sequences, as well as surrounding low copy regions, will be produced based on the information obtained in Task 2. They will be hybridized with mRNA samples that comprise developmental stages and organs representative of the wheat plant development cycle as well as with genomic DNA from sorted group 3 chromosomes and aneuploid lines to evaluate for each gene the copy number on the 3 homoeologous groups. The same array will then be hybridized with genomic DNA from a wheat diversity panel to analyze intervarietal copy number variations. T3 will be led by the INRA Clermont-Ferrand (Partner 1)
- T4, led by URGI (Partner 3), will integrate the 3B Sequence and all relevant annotations to the URGI Information System . This will allow the dissemination of the information to the international community and link the sequence to other databases to ensure a user friendly connection between the sequence, the physical map, and the genetic/traits (QTL) maps as well as with expression and SNP data. Moreover, the URGI Information System will provide online tools to support the annotation of genes and transposable elements. The same database will host data generated in the framework of the GAIN-SPEED companion project to ensure a perfect integration of resources.

